Dr. Ben Sikes, Associate Professor of Ecology and Evolutionary Biology at the University of Kansas and MAPS team lead for the soil and plant groups, has embarked on developing and teaching a follow-up collaborative, multi-university course for graduate students associated with and interested in the MAPS research. The first MAPS Biology Topics collaborative, multi-university course launched in 2018 and produced a student and faculty co-authored paper appearing in the peer-reviewed journal of the American Institute of Biological Sciences, BioScience, July 2020 issue titled Connections and Feedback: Aquatic, Plant, and Soil Microbiomes in Heterogeneous and Changing Environments. The goal of the 2018 class was to review and synthesize existing information on linkages between and feedbacks among plant, soil, and freshwater microbiomes. According to Sikes, the new course for the fall of 2020 titled Topics in Structural Equation Modeling (SEM) for Biologists seemed like a logical next step for MAPS students. This new course is a 3-credit hour Special Topics course for graduate students and is funded by the Kansas NSF EPSCoR RII Track-1 Award OIA-1656006: Microbiomes of Aquatic, Plant, and Soil Systems across Kansas (MAPS). The MAPS funding includes support for a teaching assistant and future publication efforts. Jacob Hopkins, a 4th year Ph.D. student in the Sikes Microbial Lab will serve as the teaching assistant and co-instructor for the course. Jacob is also participating in the Synthesis Team’s work on the MAPS SEM model.
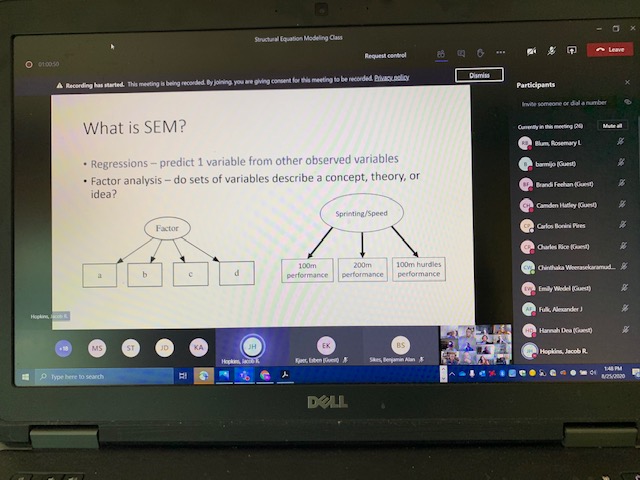
Structural Equation Modeling (SEM), a multivariate statistical analysis technique, uses two or more structural [cause-effect] equations to model and analyze multivariate relationships in which researchers can estimate multiple and interrelated dependence in a single analysis. As specified on the syllabus, SEM “allows for an intuitive graphical representation of complex networks of relationships. Complex relationships are common in STEM fields from molecular scales (gene pathways) to entire ecosystems, and often include direct and indirect relationships, mediation, and explanatory variables that are correlated. SEM can be used to represent and test hypothesized cause-effect pathways, as well as “include unobserved ’latent’ variables or pool effects of multiple variables into ‘composite’ variables. When done well, SEM allows users to test multivariate cause-effect relationships and holistically understand complex processes. SEM can also help develop new hypotheses to be targeted with experiments.”
The course description is as follows:
The COVID-19 situation and the multi-institutional nature of this course means it will be mostly online. This is an ideal course for learning virtually. Assignments are activities focused on applying information from lessons (synchronous and recorded) and readings to learn SEM. These exercises include both stepwise learning activities and a student-centered project later in the semester that integrates all the steps. All activities will use the freely available software R and can be done without extensive in-person contact. The course is set up as 3 credits with class to be held TTh, 1:00-2:15 PM. The course is mainly for graduate students, but open to upper-level undergraduates with approval from local University instructor(s). Class will be using Microsoft TEAMS.
One of the course goals is for students to gain an effective understanding of how to use SEM correctly, without having to focus on the underlying math used for SEM. In addition, students do not need to have a proficiency R code, a coding language and environment used for statistical computing and graphics, to succeed in this course as the objectives are focused only on building the R coding tools needed to do SEM. In an effort to tailor the course to each student’s specific interest, prior to the start of the course, students were asked to find a primary research paper in their field/interest area that uses SEM. These papers will be one tool to help each student utilize SEM for their own research project’s application, data types, and questions. To complete their final individual SEM project, students will be encouraged to use their own data or data from their lab groups. Discipline-specific data will help students learn and understand the idiosyncrasies of sample sizes and data structure as well as the SEM solutions associated with their field.
As of the writing of this article, there were a total of 19 students enrolled: 6 from KU Ecology and Evolutionary Biology, 6 from KSU Biology; 3 from KSU Agronomy; 3 from KSU Geology, and 1 from WSU Biology. . Faculty from KU, KSU, WSU and FHSU are participating as well.
